Research across global academic and commercial labs to create a more efficient drug discovery process won recognition today with a special Gordon Bell Prize for work fighting COVID-19.
A team of 27 researchers led by Rommie Amaro at the University of California at San Diego (UCSD) combined high performance computing (HPC) and AI to provide the clearest view to date of the coronavirus, winning the award.
Their work began in late March when Amaro lit up Twitter with a picture of part of a simulated SARS-CoV-2 virus that looked like an upside-down Christmas tree.
Seeing it, one remote researcher noticed how a protein seemed to reach like a crooked finger from behind a protective shield to touch a healthy human cell.
“I said, ‘holy crap, that’s crazy’… only through sharing a simulation like this with the community could you see for the first time how the virus can only strike when it’s in an open position,” said Amaro, who leads a team of biochemists and computer experts at UCSD.
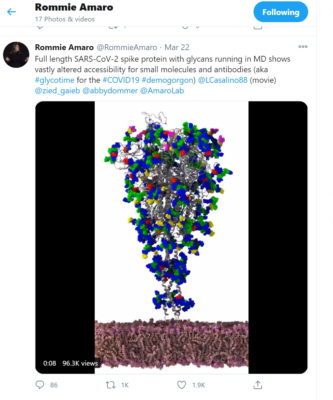
The image in the tweet was taken by Amaro’s lab using what some call a computational microscope, a digital tool that links the power of HPC simulations with AI to see details beyond the capabilities of conventional instruments.
It’s one example of work around the world using AI and data analytics, accelerated by NVIDIA Clara Discovery, to slash the $2 billion in costs and ten-year time span it typically takes to bring a new drug to market.
A Virtual Microscope Enhanced with AI
In early October, Amaro’s team completed a series of more ambitious HPC+AI simulations. They showed for the first time fine details of how the spike protein moved, opened and contacted a healthy cell.
One simulation (below) packed a whopping 305 million atoms, more than twice the size of any prior simulation in molecular dynamics. It required AI and all 27,648 NVIDIA GPUs on the Summit supercomputer at Oak Ridge National Laboratory.
More than 4,000 researchers worldwide have downloaded the results that one called “critical for vaccine design” for COVID and future pathogens.
Today, it won a special Gordon Bell Prize for COVID-19, the equivalent of a Nobel Prize in the supercomputing community.
Two other teams also used NVIDIA technologies in work selected as finalists in the COVID-19 competition created by the ACM, a professional group representing more than 100,000 computing experts worldwide.
And the traditional Gordon Bell Prize went to a team from Beijing, Berkeley and Princeton that set a new milestone in molecular dynamics, also using a combination of HPC+AI on Summit.
An AI Funnel Catches Promising Drugs
Seeing how the infection process works is one of a string of pearls that scientists around the world are gathering into a new AI-assisted drug discovery process.
Another is screening from a vast field of 1068 candidates the right compounds to arrest a virus. In a paper from part of the team behind Amaro’s work, researchers described a new AI workflow that in less than five months filtered 4.2 billion compounds down to the 40 most promising ones that are now in advanced testing.
“We were so happy to get these results because people are dying and we need to address that with a new baseline that shows what you can get with AI,” said Arvind Ramanathan, a computational biologist at Argonne National Laboratory.
Ramanathan’s team was part of an international collaboration among eight universities and supercomputer centers, each contributing unique tools to process nearly 60 terabytes of data from 21 open datasets. It fueled a set of interlocking simulations and AI predictions that ran across 160 NVIDIA A100 Tensor Core GPUs on Argonne’s Theta system with massive AI inference runs using NVIDIA TensorRT on the many more GPUs on Summit.
Docking Compounds, Proteins on a Supercomputer
Earlier this year, Ada Sedova put a pearl on the string for protein docking (described in the video below) when she described plans to test a billion drug compounds against two coronavirus spike proteins in less than 24 hours using the GPUs on Summit. Her team’s work cut to just 21 hours the work that used to take 51 days, a 58x speedup.
In a related effort, colleagues at Oak Ridge used NVIDIA RAPIDS and BlazingSQL to accelerate by an order of magnitude data analytics on results like Sedova produced.
Among the other Gordon Bell finalists, Lawrence Livermore researchers used GPUs on the Sierra supercomputer to slash the training time for an AI model used to speed drug discovery from a day to just 23 minutes.
From the Lab to the Clinic
The Gordon Bell finalists are among more than 90 research efforts in a supercomputing collaboration using 50,000 GPU cores to fight the coronavirus.
They make up one front in a global war on COVID that also includes companies such as Oxford Nanopore Technologies, a genomics specialist using NVIDIA’s CUDA software to accelerate its work.
Oxford Nanopore won approval from European regulators last month for a novel system the size of a desktop printer that can be used with minimal training to perform thousands of COVID tests in a single day. Scientists worldwide have used its handheld sequencing devices to understand the transmission of the virus.
Relay Therapeutics uses NVIDIA GPUs and software to simulate with machine learning how proteins move, opening up new directions in the drug discovery process. In September, it started its first human trial of a molecule inhibitor to treat cancer.
Startup Structura uses CUDA on NVIDIA GPUs to analyze initial images of pathogens to quickly determine their 3D atomic structure, another key step in drug discovery. It’s a member of the NVIDIA Inception program, which gives startups in AI access to the latest GPU-accelerated technologies and market partners.
From Clara Discovery to Cambridge-1
NVIDIA Clara Discovery delivers a framework with AI models, GPU-optimized code and applications to accelerate every stage in the drug discovery pipeline. It provides speedups of 6-30x across jobs in genomics, protein structure prediction, virtual screening, docking, molecular simulation, imaging and natural-language processing that are all part of the drug discovery process.
It’s NVIDIA’s latest contribution to fighting the SARS-CoV-2 and future pathogens.
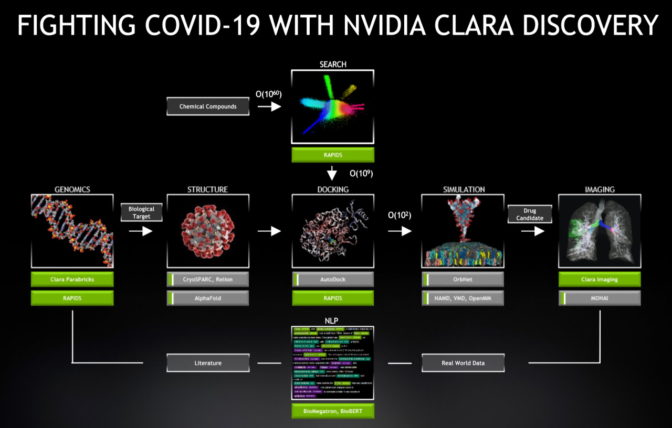
Within hours of the shelter-at-home order in the U.S., NVIDIA gave researchers free access to a test drive of Parabricks, our genomic sequencing software. Since then, we’ve provided as part of NVIDIA Clara open access to AI models co-developed with the U.S. National Institutes of Health.
We’ve also committed to build with partners including GSK and AstraZeneca Europe’s largest supercomputer dedicated to driving drug discovery forward. Cambridge-1 will be an NVIDIA DGX SuperPOD system capable of delivering more than 400 petaflops of AI performance.
Next Up: A Billion-Atom Simulation
The work is just getting started.
Ramanathan of Argonne sees a future where self-driving labs learn what experiments they should launch next, like autonomous vehicles finding their own way forward.
“And I want to scale to the absolute max of screening 1068 drug compounds, but even covering half that will be significantly harder than what we’ve done so far,” he said.
“For me, simulating a virus with a billion atoms is the next peak, and we know we will get there in 2021,” said Amaro. “Longer term, we need to learn how to use AI even more effectively to deal with coronavirus mutations and other emerging pathogens that could be even worse,” she added.
Hear NVIDIA CEO Jensen Huang describe in the video below how AI in Clara Discovery is advancing drug discovery.
At top: An image of the SARS-CoV-2 virus based on the Amaro lab’s simulation showing 305 million atoms.